|
Examples / GARCHIMPORT.RPF |
GARCHIMPORT.RPF is an example of importance sampling, applied to Monte Carlo integration on a GARCH model.
The log likelihood for a GARCH model has a non-standard form, so it isn’t possible to draw directly from the posterior distribution, even with “flat” priors. Importance sampling is one way to conduct a Bayesian analysis of such a model. An alternative is shown in GARCHGIBBS.RPF.
The importance function used here is a multivariate Student-t, with the mean being the maximum likelihood estimate and the covariance matrix being the estimated covariance matrix from GARCH. This is fattened up by using 5 degrees of freedom. (Note that the GARCH model itself is being estimated assuming Normal residuals. The fat-tailed t is used only for the importance function).
The true density function is computed using GARCH with an input set of coefficients and METHOD=EVAL which does a single function evaluation at the initial guess values. Most instructions which might be used in this way (BOXJENK, CVMODEL, FIND, MAXIMIZE, NLLS) have METHOD=EVAL options.
The example uses a GARCH model on the US 3-month Treasury Bill rate. The “mean model” is a one lag autoregression. This estimates a linear regression to get its residual variance for use as the pre-sample value throughout the analysis.
linreg ftbs3
# constant ftbs3{1}
compute h0=%sigmasq
Estimate a GARCH with Normally distributed errors
garch(p=1,q=1,reg,presample=h0,distrib=normal) / ftbs3
# constant ftbs3{1}
Set up for importance sampling.
FXX is the factor of the covariance matrix of coefficients
XBASE is the estimated coefficient vector
FBASE is the log likelihood (thus the function value at XBASE).
compute fxx =%decomp(%xx)
compute xbase=%beta
compute fbase=%logl
This is the desired number of draws and the degrees of freedom of the multivariate \(t\) distribution from which we’re drawing.
compute ndraws=10000
compute drawdf=5.0
Initialize vectors for the first and second moments of the coefficients
compute sumwt=0.0, sumwt2=0.0
compute [vect] b = %zeros(%nreg,1)
compute [symm] bxx = %zeros(%nreg,%nreg)
dec vect u(%nreg) betau(%nreg)
This is used for an estimated density of the persistence measure (ALPHA+BETA). We need to keep the values of the draws and the observation weights in order to use the DENSITY instruction.
set persist 1 ndraws = 0.0
set weights 1 ndraws = 0.0
The pseudo-code for the draw loop is
do draw=1,ndraws
...draw a random set of coefficients from a multivariate t with mean XBASE, scale covariance matrix with factor FXX and DRAWDF degrees of freedom.
...evaluate the GARCH log likelihood and compute the weight
...save the weight on the draw and do whatever other bookeeping is required
end do draw
The random set of coefficients is done easily with the help of the %RANMVT function:
compute betau=xbase+%ranmvt(fxx,drawdf)
As a side effect, %RANMVT fills in the value that can be obtained with %RANLOGKERNEL, which will have the log of the density at the draw relative to the density at the mean (XBASE):
compute gx = %ranlogkernel()
GARCH with EVAL and INIT=BETAU (the drawn set of coefficients) is used to compute the log likelihood:
garch(p=1,q=1,reg,presamp=h0,method=eval,init=betau) / ftbs3
# constant ftbs3{1}
We then need to compute the difference between the log likelihood at the draw and the log likelihood at the mode:
compute fx = %logl-fbase
The one complication is that it is possible for %LOGL to be NA in the GARCH model (if the GARCH parameters go sufficiently explosive). If it is, make the weight zero. Otherwise, exp(...) the difference in the log densities to get the weight.
compute weight = %if(%valid(%logl),exp(fx-gx),0.0)
For the bookkeeping, we are saving the (weighted) sums of the first and second moments of the coefficient vector. We are also saving the full draw history of the persistence measure (which is the sum of the 4th and 5th elements of the BETAU).
compute sumwt = sumwt+weight, sumwt2 = sumwt2+weight^2
compute b = b+weight*betau, bxx = bxx+weight*%outerxx(betau)
compute persist(draw) = betau(4)+betau(5)
compute weights(draw) = weight
Processing the Results
The efficacy of importance sampling depends upon function being estimated, but the following is a simple estimate of the number of effective draws.
?"Effective Sample Size" sumwt^2/sumwt2
Transform the accumulated first and second moments into mean and variance
compute b = b/sumwt, bxx = bxx/sumwt-%outerxx(b)
Create a table with the Monte Carlo means and standard deviations
report(action=define)
report(atrow=1,atcol=1,fillby=cols) "a" "b" "gamma" "alpha" "beta"
report(atrow=1,atcol=2,fillby=cols) b
report(atrow=1,atcol=3,fillby=cols) %sqrt(%xdiag(bxx))
report(action=show)
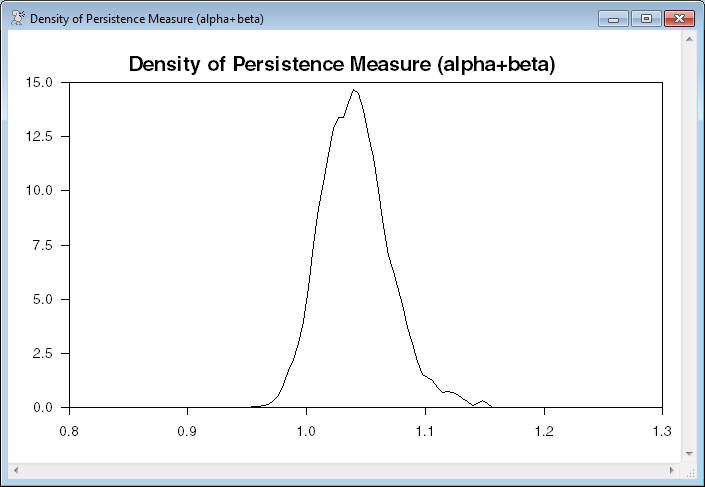
Estimate the density of the persistence measure and graph it.
density(weights=weights,smpl=weights>0.00,smoothing=1.5) $
persist 1 ndraws xx dx
scatter(style=line,header=$
"Density of Persistence Measure (alpha+beta)")
# xx dx
Full Program
open data haversample.rat
calendar(m) 1957
data(format=rats) 1957:1 2006:12 ftbs3
*
graph(header="US 3-Month Treasury Bill Rate")
# ftbs3
*
* Estimate a linear regression to get its residual variance for use as
* the pre-sample value.
*
linreg ftbs3
# constant ftbs3{1}
compute h0=%sigmasq
*
* Estimate a GARCH with Normally distributed errors
*
garch(p=1,q=1,reg,presample=h0,distrib=normal) / ftbs3
# constant ftbs3{1}
compute normallogl=%logl
*
* Set up for importance sampling.
* FXX is the factor of the covariance matrix of coefficients
* XBASE is the estimated coefficient vector
* FBASE is the final log likelihood
*
compute fxx =%decomp(%xx)
compute xbase=%beta
compute fbase=%logl
*
* This is the number of draws and the degrees of freedom of the
* multivariate t distribution from which we're drawing.
*
compute ndraws=10000
compute drawdf=5.0
*
* Initialize vectors for the first and second moments of the coefficients
*
compute sumwt=0.0,sumwt2=0.0
compute [vect] b=%zeros(%nreg,1)
compute [symm] bxx=%zeros(%nreg,%nreg)
dec vect u(%nreg) betau(%nreg)
*
* This is used for an estimated density of the persistence measure
* (alpha+beta). We need to keep the values of the draws and the
* observation weights in order to use the density instruction.
*
set persist 1 ndraws = 0.0
set weights 1 ndraws = 0.0
*
do draw=1,ndraws
*
* Draw coefficients from a multivariate t
*
compute betau=xbase+%ranmvt(fxx,drawdf)
*
* Compute the density of the draw relative to the density at the
* mode. The log of this is returned by the value of %ranlogkernel()
* from the %ranmvt function.
*
compute gx=%ranlogkernel()
*
* Compute the log likelihood at the drawn value of the coefficients
*
garch(p=1,q=1,reg,presample=h0,method=eval,initial=betau) / ftbs3
# constant ftbs3{1}
*
* Compute the difference between the log likelihood at the draw and
* the log likelihood at the mode.
*
compute fx=%logl-fbase
*
* It's quite possible for %logl to be NA in the GARCH model (if the
* GARCH parameters go sufficiently explosive). If it is, make the
* weight zero. Otherwise exp(...) the difference in the log densities.
*
compute weight=%if(%valid(%logl),exp(fx-gx),0.0)
*
* Update the accumulators. These are weighted sums.
*
compute sumwt=sumwt+weight,sumwt2=sumwt2+weight^2
compute b=b+weight*betau
compute bxx=bxx+weight*%outerxx(betau)
*
* Save the drawn value for persist and weight.
*
compute persist(draw)=betau(4)+betau(5)
compute weights(draw)=weight
end do draws
*
* The efficacy of importance sampling depends upon function being
* estimated, but the following is a simple estimate of the number of
* effective draws.
*
disp "Effective Sample Size" sumwt^2/sumwt2
*
* Transform the accumulated first and second moments into mean and
* variance.
*
compute b=b/sumwt,bxx=bxx/sumwt-%outerxx(b)
*
* Create a table with the Monte Carlo means and standard deviations
*
report(action=define)
report(atrow=1,atcol=1,fillby=cols) "a" "b" "gamma" "alpha" "beta"
report(atrow=1,atcol=2,fillby=cols) b
report(atrow=1,atcol=3,fillby=cols) %sqrt(%xdiag(bxx))
report(action=show)
*
* Estimate the density of the persistence measure and graph it
*
density(weights=weights,smpl=weights>0.00,smoothing=1.5) $
persist 1 ndraws xx dx
scatter(style=line,header=$
"Density of Persistence Measure (alpha+beta)")
# xx dx
Output
Linear Regression - Estimation by Least Squares
Dependent Variable FTBS3
Monthly Data From 1957:02 To 2006:12
Usable Observations 599
Degrees of Freedom 597
Centered R^2 0.9719842
R-Bar^2 0.9719373
Uncentered R^2 0.9941920
Mean of Dependent Variable 5.3828881469
Std Error of Dependent Variable 2.7551056598
Standard Error of Estimate 0.4615335100
Sum of Squared Residuals 127.16886894
Regression F(1,597) 20712.4001
Significance Level of F 0.0000000
Log Likelihood -385.7953
Durbin-Watson Statistic 1.3186
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. Constant 0.0816496008 0.0413816505 1.97309 0.04894649
2. FTBS3{1} 0.9853633822 0.0068466985 143.91803 0.00000000
GARCH Model - Estimation by BFGS
Convergence in 38 Iterations. Final criterion was 0.0000046 <= 0.0000100
Dependent Variable FTBS3
Monthly Data From 1957:02 To 2006:12
Usable Observations 599
Log Likelihood -61.3428
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. Constant 0.0631109748 0.0228117343 2.76660 0.00566440
2. FTBS3{1} 0.9900702410 0.0051585759 191.92705 0.00000000
3. C 0.0014556715 0.0005207479 2.79535 0.00518439
4. A 0.3065910350 0.0432686471 7.08576 0.00000000
5. B 0.7300816921 0.0297683170 24.52546 0.00000000
The remainder is based upon random numbers, and so won't match exactly if you run it.
Effective Sample Size 2993.40310
a 0.062580 0.021771
b 0.990223 0.004923
gamma 0.001834 0.000716
alpha 0.327963 0.051931
beta 0.713289 0.035107
Graphs
Copyright © 2026 Thomas A. Doan