|
Examples / VECMGARCH.RPF |
VECMGARCH.RPF is an example of some of the calculations done in Pardo and Torro(2007). This is applied to a different data set than in the paper; in this case, the closing weekly adjusted values for the Dow Jones Industrial Average and the Russell 2000 for the U.S. stock market. Note that, unlike most applications of GARCH to stock market data, these are not done in returns—if you convert to returns, you would lose the (possible) cointegration, and analyzing this as an error-correction GARCH model is the whole point. The two series are transformed to 100*log's and are normalized to both start at 0. The normalization has no effect on the model, but makes it easier to see (graphically) that the two series might be cointegrated.
set logdjia = 100.0*(log(djia/djia(1)))
set logrut = 100.0*(log(russell2000/russell2000(1)))
AIC and SBC (and HQ) pick very different lag lengths. The SBC picks 1, which seems too short. We use 4 in the subsequent analysis, which is a compromise which is at least a local minimum in the AIC. Note that the lag selection criteria assume homoscedasticity, and so can be thrown off by GARCH error processes.
There's no strong reason to believe that the two stock indexes are cointegrated since they have different risk-return structures. The Johansen test allowing for trending data is done with @JOHMLE. The Johansen test is known to be at least somewhat robust to a GARCH error process.
@johmle(lags=nlags,det=constant,cv=cv)
# logdjia logrut
The Johansen test marginally rejects cointegration, but we'll go ahead and assign cointegration rank one for illustrative purposes. This uses the estimated cointegrating vector. In other applications (for instance with spot vs futures prices), the cointegrating vector will be known theoretically, and you should use that, not an estimated one.
This sets up a VECM for the DET=CONSTANT model (including a CONSTANT as a DET term in the VAR). For this, you put in the original dependent variables, not the differences. The rearrangement of the regression is done internally when you ESTIMATE the model.
equation(coeffs=cv) ecteq *
# logdjia logrut
*
system(model=basevecm)
variables logdjia logrut
lags 1 to nlags
det constant
ect ecteq
end(system)
The error correction model is first estimated by least squares, and a test for ARCH is done on the residuals with @MVARCHTEST, which overwhelmingly rejects homoscedasticity.
estimate(resids=vecmresids)
@mvarchtest(lags=5)
# vecmresids
This estimates an asymmetric BEKK GARCH model. This saves the jointly standardized residuals (as STDU), the conditional covariance matrices (HH) and (non-standardized) residuals (RR) and the VECH representation (VECHCOMPS):
garch(model=basevecm,mv=bekk,asymmetric,stdresids=stdu,hmatrices=hh,rvectors=rr,$
pmethod=simplex,piters=10,method=bfgs,iters=500,vechmat=vechcomps)
One thing to note in the output is that neither of the error correction terms is significant. If the series were truly cointegrated (again, we are doing this as a VECM for illustration) at least one of these two should be significant. These t-statistics are the tests for long-run causality.
This does the recommended set of multivariate residual diagnostics (for remaining ARCH and for serial correlation in the mean):
@mvarchtest(lags=5)
# stdu
@mvqstat(lags=5)
# stdu
The results pass easily.
These next lines do causality tests in the mean. The first does a joint test for whether the dx2 lags and the ect lag are zero in the first equation (first block of mean model coefficients) and the second does a similar test for the dx1 lags and ect in the second block of mean model coefficients. The simplest way to set these up is to use the Regression Tests, Exclusion Restrictions test wizard. Note that the Pardo-Torro paper doesn't include the lagged ECT in the test, but it should be included since if it's non-zero, the lagged "other" does help to predict. The wizard dialog for the x2 to x1 causality will look something like
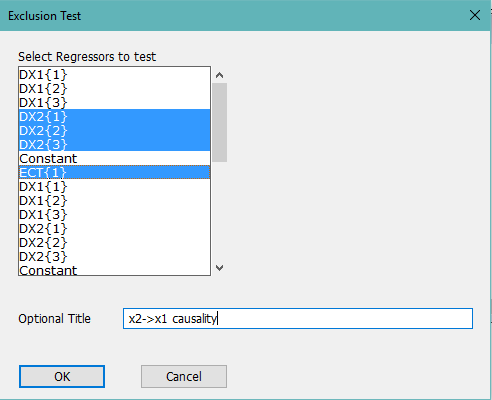
The first block of regressors is for the first equation, so this will do a joint test on the DX2 lags and the ECT lag for that.
test(zeros,title="x2-->x1 causality")
# 4 5 6 8
test(zeros,title="x1-->x2 causality")
# 9 10 11 16
Neither direction is significant. There are also tests on the BEKK cross effects (joint test of all the off-diagonal A, B and D coefficients) and a joint test on all the asymmetry (D) coefficients. Again, it's easiest to set these up using the Regression Tests wizard.
There are also diagnostics on the univariate standardized residuals:
set std1 = rr(t)(1)/sqrt(hh(t)(1,1))
set std2 = rr(t)(2)/sqrt(hh(t)(2,2))
You can only do univariate tests (not joint ones like @MVARCHTEST) on these. Again, these pass all the tests. However, note well that we chose to fit this under the assumption of cointegration despite the fact that the initial test showed that to be likely incorrect—the model is probably more complicated than it needs to be to adequately explain the data.
None of the remaining analysis has anything to do with this being a VECM-GARCH model—they're general to the multivariate GARCH model. Graphs are done of the conditional volatility (in standard deviations, which aren't dominated by the more extreme values as the variances), the conditional correlation and the conditional beta. All of these can be done (in pretty much the same way) for any type of multivariate GARCH model.
The final calculations are impact surfaces for the two variances and the covariance with respect to lagged residuals. This can be computed (easily) for any multivariate GARCH model which has a VECH representation (DVECH, VECH or any form of BEKK), but the graphs are specific to this being a two-variable model, since it does a contour graph with axes for each shock.
Full Program
open data "usstocks.xls"
calendar(w) 2006:1:2
*
* The data were taken from Yahoo Finance and are in reversed
* chronological order. RATS v9.1 automatically detects that and
* rearranges the data. (The REVERSE option has no effect, but creates an
* immediate error with earlier versions of RATS).
*
data(format=xls,org=columns,reverse) / djia russell2000
*
* Normalize both to 100 * log relative to the first entry
*
set logdjia = 100.0*(log(djia/djia(1)))
set logrut = 100.0*(log(russell2000/russell2000(1)))
*
graph(footer="Log Stock Indexes",key=upleft,klabels=||"DJIA","Russell 2000"||) 2
# logdjia
# logrut
*
* AIC and SBC (or HQ) pick very different lag lengths. The 1 favored by
* SBC and HQ seems too short. AIC has a local minimum at 4, so we choose
* that. (None of these criteria are designed for heteroscedastic data,
* so they can be thrown off by outliers).
*
@varlagselect(lags=10,crit=aic)
# logdjia logrut
@varlagselect(lags=10,crit=sbc)
# logdjia logrut
*
compute nlags=4
*
* There's no strong reason to believe that the two stock indexes are
* cointegrated since they have different risk-return structures. The
* Johansen test marginally rejects cointegration, but we'll go ahead and
* assign cointegration rank one for illustrative purposes. This uses the
* estimated cointegrating vector. In other applications (for instance
* with spot vs futures prices), the cointegrating vector will be known
* theoretically, and you should use that, not an estimated one.
*
* This analyzes cointegration using DET=CONSTANT, which allows for a
* trend in the series.
*
@johmle(lags=nlags,det=constant,cv=cv)
# logdjia logrut
*
* This sets up a VECM for the DET=CONSTANT model (including a CONSTANT
* as a DET term in the VAR). For this, you put in the original dependent
* variables, not the differences. The rearrangement of the regression is
* done internally when you ESTIMATE the model.
*
equation(coeffs=cv) ecteq *
# logdjia logrut
*
system(model=basevecm)
variables logdjia logrut
lags 1 to nlags
det constant
ect ecteq
end(system)
*
estimate(resids=vecmresids)
*
* Note that the number of lags used in the diagnostic tests has nothing
* to do with the choice of the number of lags in the VAR model. (The
* paper does some tests later on the OLS VECM residuals and uses 5 for
* those). Somewhere in the 3-5 lag range generally seems appropriate for
* this.
*
@mvarchtest(lags=5)
# vecmresids
*
* Estimate an asymmetric BEKK GARCH model. This saves the jointly
* standardized residuals (as STDU), the conditional covariance matrices
* (HH) and (non-standardized) residuals (RR) and the VECH representation
* (VECHCOMPS).
*
garch(model=basevecm,mv=bekk,asymmetric,stdresids=stdu,hmatrices=hh,rvectors=rr,$
pmethod=simplex,piters=10,method=bfgs,iters=500,vechmat=vechcomps)
*
* These are (our) recommended diagnostic tests on the jointly standardized
* residuals. Tests on the univariate standardized residuals come later.
*
@mvarchtest(lags=5)
# stdu
@mvqstat(lags=5)
# stdu
*
* Causality tests in the mean. The first tests does a joint test for
* whether the dx2 lags and the ect lag are zero in the first equation
* (first block of mean model coefficients) and the second does a similar
* test for the dx1 lags and ect in the second block of mean model
* coefficients. (You can use the Statistics-Regression Tests, Exclusion
* Restrictions wizard to set these up.)
*
* (The paper doesn't include the ECT in the test, but it should be
* included since if it's non-zero, the lagged "other" does help to
* predict).
*
test(zeros,title="x2-->x1 causality")
# 4 5 6 8
test(zeros,title="x1-->x2 causality")
# 9 10 11 16
*
* Wald tests of BEKK coefficients
*
test(zeros,title="BEKK cross effects")
# 21 22 25 26 29 30
test(zeros,title="Asymmetry")
# 28 29 30 31
*
* Tests on the univariate standardized residuals
*
set std1 = rr(t)(1)/sqrt(hh(t)(1,1))
set std2 = rr(t)(2)/sqrt(hh(t)(2,2))
*
* Skewness, (excess) kurtosis and normality (Jarque-Bera) are all part
* of the standard STATISTICS output.
*
stats std1
stats std2
*
* The Q(20) (Ljung-Box) and Q^2(20) (McLeod-Li) are included in the
* BDINDTESTS procedure.
*
@bdindtests(number=20) std1
@bdindtests(number=20) std2
*
* You can also do the LB and McLeod-Li tests separately using @RegCorrs
* (for LB) and @McLeodLi. The results are very slightly different
* because they use a slightly different calculation for the
* autocorrelations.
*
@regcorrs(qstats,number=20) std1
@mcleodli(number=20) std1
@archtest(lags=20) std1
*
@regcorrs(qstats,number=20) std2
@mcleodli(number=20) std2
@archtest(lags=20) std2
*
* These graph the conditional volatility (in standard deviations to
* allow more detail to be seen), the conditional correlation and the
* conditional beta. The latter two also include a horizontal line at the
* corresponding value from the original VECM estimates.
*
set sig11 = sqrt(hh(t)(1,1))
set sig22 = sqrt(hh(t)(2,2))
graph(footer="Conditional Volatility in Std Deviations",$
key=below,klabels=||"DJIA","Russell 2000"||) 2
# sig11
# sig22
*
set rho12 = %cvtocorr(hh(t))(1,2)
graph(footer="Conditional correlation",vgrid=%cvtocorr(%sigma)(1,2))
# rho12
*
set beta12 = hh(t)(1,2)/hh(t)(1,1)
graph(footer="Conditional beta of Russell 2000 with respect to DJIA",$
vgrid=%sigma(1,2)/%sigma(1,1))
# beta12
*
* Compute impact surfaces (done as contour graphs)
*
compute ngrid=21
*
* This picks the grids in each direction based upon the observed
* absolute values of the residuals from the VECM (going from - to + of
* the 99%-ile of the absolute value).
*
set plus1 = abs(vecmresids(1))
stats(fract,noprint) plus1
compute grid1=%seqrange(-%fract99,+%fract99,ngrid)
set plus2 = abs(vecmresids(2))
stats(fract,noprint) plus2
compute grid2=%seqrange(-%fract99,+%fract99,ngrid)
*
dec rect var1g(ngrid,ngrid) var2g(ngrid,ngrid) covarg(ngrid,ngrid)
*
do i=1,ngrid
do j=1,ngrid
compute [symm] response=%vectosymm($
vechcomps("A")*%vec(%outerxx(||grid1(i)|grid2(j)||))+$
vechcomps("D")*%vec(%outerxx(||%minus(grid1(i))|%minus(grid2(j))||)),$
2)
compute var1g(i,j)=response(1,1)
compute var2g(i,j)=response(2,2)
compute covarg(i,j)=response(1,2)
end do j
end do i
*
* Do contour plots
*
gcontour(x=grid1,y=grid2,f=var1g,hlabel="DJIA",vlabel="Russell 2000",$
footer="Impact Surface for DJIA Variance")
gcontour(x=grid1,y=grid2,f=var2g,hlabel="DJIA",vlabel="Russell 2000",$
footer="Impact Surface for Russell 2000 Variance")
gcontour(x=grid1,y=grid2,f=covarg,hlabel="DJIA",vlabel="Russell 2000",$
footer="Impact Surface for Covariance")
Output
VAR Lag Selection
Lags AICC
0 15.9792555
1 8.2352485
2 8.2361942
3 8.2354481
4 8.2272985
5 8.2287103
6 8.2358474
7 8.2135519*
8 8.2143388
9 8.2213291
10 8.2286912
VAR Lag Selection
Lags SBC/BIC
0 15.9915266
1 8.2720192*
2 8.2974074
3 8.3210460
4 8.3372231
5 8.3629029
6 8.3942490
7 8.3961029
8 8.4209793
9 8.4519985
10 8.4833285
Likelihood Based Analysis of Cointegration
Variables: LOGDJIA LOGRUT
Estimated from 2006:01:30 to 2020:08:10
Data Points 759 Lags 4 with Constant
Unrestricted eigenvalues, -T log(1-lambda) and Trace Test
Roots Rank EigVal Lambda-max Trace Trace-95%
2 0 0.0125 9.5275 10.3932 15.3400
1 1 0.0011 0.8657 0.8657 3.8400
Cointegrating Vector for Largest Eigenvalue
LOGDJIA LOGRUT
-0.168654 0.130579
VAR/System - Estimation by Cointegrated Least Squares
Weekly Data From 2006:01:30 To 2020:08:10
Usable Observations 759
Dependent Variable LOGDJIA
Mean of Dependent Variable 0.0828052299
Std Error of Dependent Variable 2.3021718535
Standard Error of Estimate 2.2865403259
Sum of Squared Residuals 3926.4282631
Durbin-Watson Statistic 2.0046
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. D_LOGDJIA{1} -0.049703854 0.071699459 -0.69322 0.48838272
2. D_LOGDJIA{2} 0.119169085 0.071973693 1.65573 0.09819403
3. D_LOGDJIA{3} -0.061265243 0.071875202 -0.85238 0.39427297
4. D_LOGRUT{1} 0.013719874 0.053791745 0.25506 0.79875015
5. D_LOGRUT{2} -0.059179461 0.053736351 -1.10129 0.27112204
6. D_LOGRUT{3} -0.049054788 0.053545600 -0.91613 0.35989224
7. Constant -0.070703634 0.190250917 -0.37163 0.71027049
8. EC1{1} 0.079487651 0.082996156 0.95773 0.33850870
Dependent Variable LOGRUT
Mean of Dependent Variable 0.1237170066
Std Error of Dependent Variable 3.0781015253
Standard Error of Estimate 3.0650010017
Sum of Squared Residuals 7055.0675865
Durbin-Watson Statistic 1.9957
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. D_LOGDJIA{1} 0.068398090 0.096109791 0.71167 0.47689250
2. D_LOGDJIA{2} 0.233982630 0.096477389 2.42526 0.01553225
3. D_LOGDJIA{3} -0.086128520 0.096345367 -0.89396 0.37163181
4. D_LOGRUT{1} -0.060815734 0.072105334 -0.84343 0.39925708
5. D_LOGRUT{2} -0.115187050 0.072031080 -1.59913 0.11021235
6. D_LOGRUT{3} -0.014199461 0.071775388 -0.19783 0.84323013
7. Constant 0.278516870 0.255022510 1.09213 0.27512768
8. EC1{1} -0.072420206 0.111252488 -0.65095 0.51527557
Multivariate ARCH Test
Statistic Degrees Signif
473.79 45 0.00000
MV-GARCH, BEKK - Estimation by BFGS
Convergence in 134 Iterations. Final criterion was 0.0000000 <= 0.0000100
Weekly Data From 2006:01:30 To 2020:08:10
Usable Observations 759
Log Likelihood -2941.5485
Variable Coeff Std Error T-Stat Signif
************************************************************************************
Mean Model(LOGDJIA)
1. D_LOGDJIA{1} -0.043763647 0.060086512 -0.72834 0.46640306
2. D_LOGDJIA{2} 0.036723159 0.056000685 0.65576 0.51197683
3. D_LOGDJIA{3} -0.093426788 0.055747981 -1.67588 0.09376219
4. D_LOGRUT{1} 0.011370267 0.042423556 0.26802 0.78868564
5. D_LOGRUT{2} -0.034424126 0.041568156 -0.82814 0.40759294
6. D_LOGRUT{3} 0.039632920 0.042197520 0.93922 0.34761580
7. Constant -0.075739179 0.147442108 -0.51369 0.60747044
8. EC1{1} 0.083814979 0.060300642 1.38995 0.16454355
Mean Model(LOGRUT)
9. D_LOGDJIA{1} 0.028377503 0.081875208 0.34659 0.72889592
10. D_LOGDJIA{2} 0.087792063 0.077602752 1.13130 0.25792847
11. D_LOGDJIA{3} -0.068980642 0.078063419 -0.88365 0.37688586
12. D_LOGRUT{1} -0.043920165 0.060368653 -0.72753 0.46689973
13. D_LOGRUT{2} -0.063394688 0.059101909 -1.07263 0.28343558
14. D_LOGRUT{3} 0.027708083 0.059374618 0.46667 0.64073926
15. Constant 0.196340052 0.195734076 1.00310 0.31581459
16. EC1{1} -0.024338175 0.081203598 -0.29972 0.76439232
17. C(1,1) 0.536742145 0.087928428 6.10431 0.00000000
18. C(2,1) 1.011704111 0.133694310 7.56729 0.00000000
19. C(2,2) 0.001824514 0.134337611 0.01358 0.98916382
20. A(1,1) -0.088499362 0.130987858 -0.67563 0.49927542
21. A(1,2) -0.449294604 0.167375786 -2.68435 0.00726717
22. A(2,1) 0.107685498 0.100836352 1.06792 0.28555507
23. A(2,2) 0.356808843 0.132576854 2.69134 0.00711665
24. B(1,1) 1.053401968 0.032624545 32.28863 0.00000000
25. B(1,2) 0.150456003 0.056120300 2.68096 0.00734124
26. B(2,1) -0.155429493 0.033309666 -4.66620 0.00000307
27. B(2,2) 0.746215374 0.054142386 13.78246 0.00000000
28. D(1,1) 0.132160274 0.099141459 1.33305 0.18251622
29. D(1,2) 0.002417575 0.153900038 0.01571 0.98746676
30. D(2,1) 0.311238147 0.070389598 4.42165 0.00000980
31. D(2,2) 0.498929282 0.116488548 4.28308 0.00001843
Multivariate ARCH Test
Statistic Degrees Signif
47.81 45 0.35941
Multivariate Q Test
Test Run Over 2006:01:30 to 2020:08:10
Lags Tested 5
Degrees of Freedom 20
Q Statistic 11.06726
Signif Level 0.94446
x2-->x1 causality
Chi-Squared(4)= 3.961522 or F(4,*)= 0.99038 with Significance Level 0.41123831
x1-->x2 causality
Chi-Squared(4)= 2.229419 or F(4,*)= 0.55735 with Significance Level 0.69364712
BEKK cross effects
Chi-Squared(6)= 58.532676 or F(6,*)= 9.75545 with Significance Level 0.00000000
Asymmetry
Chi-Squared(4)= 87.878354 or F(4,*)= 21.96959 with Significance Level 0.00000000
Statistics on Series STD1
Weekly Data From 2006:01:30 To 2020:08:10
Observations 759
Sample Mean -0.002279 Variance 0.994189
Standard Error 0.997090 SE of Sample Mean 0.036192
t-Statistic (Mean=0) -0.062965 Signif Level (Mean=0) 0.949811
Skewness -0.520557 Signif Level (Sk=0) 0.000000
Kurtosis (excess) 0.667615 Signif Level (Ku=0) 0.000186
Jarque-Bera 48.374556 Signif Level (JB=0) 0.000000
Statistics on Series STD2
Weekly Data From 2006:01:30 To 2020:08:10
Observations 759
Sample Mean 0.003256 Variance 0.986803
Standard Error 0.993380 SE of Sample Mean 0.036057
t-Statistic (Mean=0) 0.090308 Signif Level (Mean=0) 0.928066
Skewness -0.462865 Signif Level (Sk=0) 0.000000
Kurtosis (excess) 0.344532 Signif Level (Ku=0) 0.053782
Jarque-Bera 30.855813 Signif Level (JB=0) 0.000000
Independence Tests for Series STD1
Test Statistic P-Value
Ljung-Box Q(20) 17.218262 0.6388
McLeod-Li(20) 18.178192 0.5757
Turning Points 1.924922 0.0542
Difference Sign -1.507874 0.1316
Rank Test 0.377956 0.7055
Independence Tests for Series STD2
Test Statistic P-Value
Ljung-Box Q(20) 10.5891603 0.9562
McLeod-Li(20) 14.1788376 0.8213
Turning Points 0.5458733 0.5852
Difference Sign 0.3769685 0.7062
Rank Test 0.2374944 0.8123
McLeod-Li Test for Series STD1
Using 759 Observations from 2006:01:30 to 2020:08:10
Test Stat Signif
McLeod-Li(20-0) 17.8764915 0.59554
Test for ARCH in STD1
Using data from 2006:01:30 to 2020:08:10
Lags Statistic Signif. Level
20 0.870 0.62672
McLeod-Li Test for Series STD2
Using 759 Observations from 2006:01:30 to 2020:08:10
Test Stat Signif
McLeod-Li(20-0) 14.0530485 0.82780
Test for ARCH in STD2
Using data from 2006:01:30 to 2020:08:10
Lags Statistic Signif. Level
20 0.697 0.83174
Graphs
This is a graph of the normalized log data
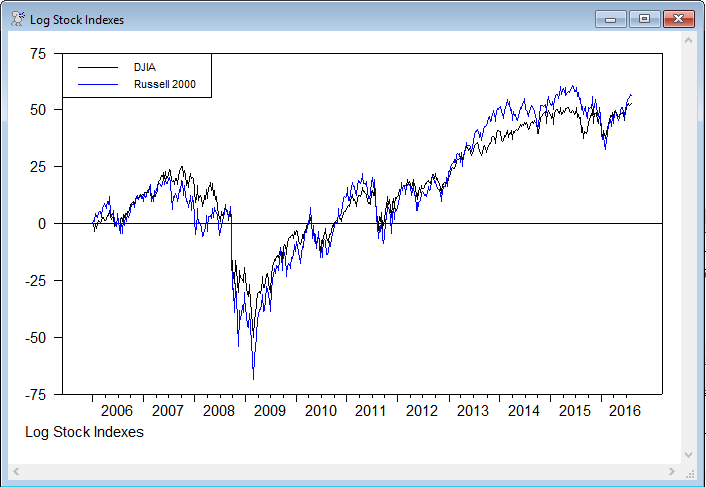 e
e
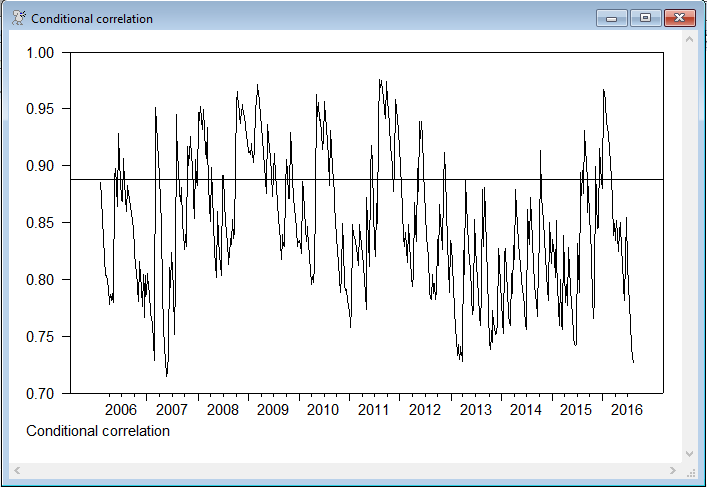
Graph of the conditional correlation with the correlation of the fixed VECM residuals shown.
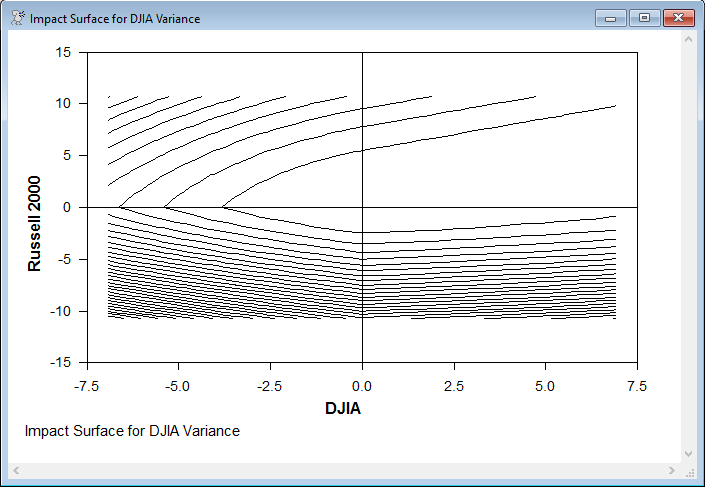
One of the news impact surfaces, graphed as a contour graph for the impact versus the shocks in the two variables. Even though this is for the variance of the DJIA, it is most heavily impacted by the negative lagged shocks in the Russell 2000 (the contours are steepest in the negative Y direction).
Copyright © 2026 Thomas A. Doan