|
Examples / MONTENEARSVAR.RPF |
MONTENEARSVAR.RPF is an example of a MCMC (Markov Chain Monte Carlo) analysis of a combination of a near VAR for the lag coefficients and a structural VAR for the covariance matrix. This is based upon the SIMSIRF.RPF example from the Sims and Zha(1999) replication files. However, this is only for illustration—it treats as exogenous a variable which really isn't, which produces some odd results.
Technically, this uses Metropolis-within-Gibbs. The structural (contemporaneous) model is drawn using Random Walk Metropolis. Given the values of the structural parameters, the variances of the shocks can be drawn directly, and given the covariance matrix, the coefficients can be drawn using the standard methods for linear SUR systems of modest size. This uses the @SURGibbsSetup procedures to handle the specialized calculations for Gibbs sampling on the near-VAR.
This same basic method can be used for an informative prior on the lag coefficients combined with a structural model for the covariance matrix. The subcalculation
compute bdraw=%ranmvposthb(hdata,hbdata)
can be replaced by
compute bdraw=%ranmvposthb(hdata+hprior,hbdata+hprior*bprior)
where HPRIOR is the precision matrix on the lag coefficients and BPRIOR is the mean.
Note that this is a very complicated sampling process which may require significant experimentation to achieve reasonable results. If you are unfamiliar with general MCMC sampling, we would recommend the RATS Bayesian Econometrics e-course. The distribution for the random walk increments for the structural models is a multivariate t governed by the following:
*
* This is the covariance matrix for the RW increment. We might have to
* change the scaling on this to achieve a reasonable acceptance
* probability.
*
compute f=%decomp(%xx)*.35
compute nudraw=6
Both the scaling (the .35) and the degrees of freedom (the NUDRAW value) may need to be changed in another application to get a reasonable rate of acceptance.
This combines the features of MONTESUR.RPF, which does Gibbs sampling for a "near-VAR", but for a simple contemporaneous model, and MONTESVAR.RPF, which does sampling for a structural contemporaneous model (SVAR) but for a full (OLS) VAR for the lag coefficients.
Full Program
open data simszha.xls
calendar(q) 1948
data(format=xls,org=columns) 1948:01 1989:03 gin82 gnp82 gnp gd lhur fygn3 m1
*
set gin82 = log(gin82)
set gnp82 = log(gnp82)
set gd = log(gd)
set m1 = log(m1)
set fygn3 = fygn3*.01
set lhur = lhur*.01
*
compute nlags =4
compute nvar =6
compute nsteps=25
compute nburn =2000
compute ndraws=10000
*
* Create a near-VAR that treats gin82 as exogenous.
*
system(model=fivevar)
variables fygn3 m1 gnp82 gd lhur
lags 1 to nlags
det constant gin82{1 to nlags}
end(system)
*
equation gineqn gin82
# constant gin82{1 to nlags}
*
system(model=nearvar)
variables fygn3 m1 gnp82 gd lhur gin82
lags 1 to nlags
det constant
end(system)
*compute nearvar=fivevar+gineqn
*
* Set up the Gibbs sampler for the SUR system
*
@SURGibbsSetup nearvar
*
* Do a SUR to get a set of estimates to initialize the Gibbs sampler.
*
sur(model=nearvar,cvout=sigmaAtBeta)
compute tnobs =%nobs
*
* Create the set of non-linear parameters, and the "A" formula for
* CVMODEL. Since these are all off-diagonal, we can try just zeros
* for the guess values.
*
nonlin(parmset=simszha,zeros) a12 a21 a23 a24 a31 a36 a41 a43 a46 a51 a53 a54 a56
compute nfree=13
*
dec frml[rect] afrml
frml afrml = ||1.0 ,a12 ,0.0 ,0.0 ,0.0 ,0.0|$
a21 ,1.0 ,a23 ,a24 ,0.0 ,0.0|$
a31 ,0.0 ,1.0 ,0.0 ,0.0 ,a36|$
a41 ,0.0 ,a43 ,1.0 ,0.0 ,a46|$
a51 ,0.0 ,a53 ,a54 ,1.0 ,a56|$
0.0 ,0.0 ,0.0 ,0.0 ,0.0 ,1.0||
*
* Compute the maximum of the log of the marginal posterior density for
* the A coefficients with a prior of the form |D|**(-delta). Delta
* should be at least (nvar+1)/2.0 to ensure an integrable posterior
* distribution. This will be used to initialize the M-H sampler and
* provide (with scaling) the covariance matrix for the increment for
* Random Walk Metropolis.
*
compute delta=3.5
cvmodel(parmset=simszha,pdf=delta,dmatrix=marginalized,method=bfgs) sigmaAtBeta afrml
*
compute shocklabels=||"MS","MD","Y","P","U","I"||
compute varlabels=||"R","M","Y","P","U","I"||
*
* This is the covariance matrix for the RW increment. We might have to
* change the scaling on this to achieve a reasonable acceptance
* probability.
*
compute f=%decomp(%xx)*.35
compute nudraw=6
*
* We need to save the draws in %%RESPONSES (for use with MCGRAPHIRF).
*
declare rect[series] impulses(nvar,nvar)
declare vect[rect] %%responses(ndraws)
*
compute thetaDraw =%parmspeek(simszha)
compute accept =0
declare vect lambdadraw(nvar) vdiag(nvar)
*
* This is for saving the draws for the structural VAR parameters.
*
dec series[vect] tgibbs
gset tgibbs 1 ndraws = thetaDraw
*
infobox(action=define,progress,lower=-nburn,upper=ndraws) $
"Metropolis within Gibbs"
do draw=-nburn,ndraws
*
* Unlike the full VAR in SIMSIRF, the likelihood for a near VAR
* doesn't isolate the structural model for the covariance matrix from
* the lag coefficients. For the full VAR, the structural coefficients
* can be drawn using only the covariance matrix from the OLS
* estimates; here, we need to use the recomputed covariance matrix at
* each Gibbs sweep. Also unlike SIMSIRF, we need to draw coefficients
* with each sweep, not just after the burn-in. (With the full VAR,
* the coefficients are needed only for doing the impulse response
* functions).
*
* So the procedure is:
*
* 1. Compute the log likelihood for the structural model given the
* covariance matrix of residuals from the current draw for the
* coefficients (<<sigmaAtBeta>>).
*
* 2. Draw a candidate set of structural parameters and compute the
* log likelihood at those.
*
* 3. Do a Metropolis acceptance test using those two to determine
* whether to reject or accept the candidate draw.
*
* 4. Draw the diagonal elements for the structural covariance matrix
* using the chosen set of structural parameters and <<sigmaAtBeta>>.
*
* 5. Draw the coefficients from the SUR, and recompute <<sigmaAtBeta>>
* for use in the next sweep.
*
* Note that there is no DFC option in the CVMODEL---that's needed only
* when the beta's can be marginalized out.
*
cvmodel(parmset=simszha,pdf=delta,dmatrix=marginalized,method=evaluate,a=afrml) sigmaAtBeta
compute logplast=%funcval
*
* Draw a new theta based at previous value
*
compute theta=thetadraw+%ranmvt(f,nudraw)
*
* Evaluate the model there
*
compute %parmspoke(simszha,theta)
cvmodel(parmset=simszha,pdf=delta,dmatrix=marginalized,method=evaluate,a=afrml) sigmaAtBeta
compute logptest=%funcval
*
* Compute the acceptance probability
*
compute alpha =exp(logptest-logplast)
if %ranflip(alpha)
compute thetadraw=theta,logplast=logptest,accept=accept+1
infobox(current=draw) %strval(100.0*accept/(draw+nburn+1),"##.#")
*
* Conditioned on theta, make a draw for lambda.
*
compute %parmspoke(simszha,thetadraw)
compute a=afrml(1)
compute vdiag =%mqformdiag(sigmaAtBeta,tr(a))
ewise lambdadraw(i)=(tnobs/2.0)*vdiag(i)/%rangamma(.5*(tnobs)+delta+1)
*
* Combine Lambda and A to generate the factor for the SVAR (for use
* in computing the impulse responses), and the draw for the
* structural precision (inverse covariance) matrix.
*
compute factor=inv(a)*%diag(%sqrt(lambdadraw))
compute hdraw=tr(a)*inv(%diag(lambdadraw))*a
*
* Compute the required information with the interaction between
* the precision matrix and the data
*
@SURGibbsDataInfo hdraw hdata hbdata
*
* Draw coefficients given the precision matrix hdraw
*
compute hpost=hdata
compute vpost=inv(hpost)
compute bpost=vpost*hbdata
compute bdraw=bpost+%ranmvnormal(%decomp(vpost))
*
* Compute covariance matrix of residuals at the current beta. (This
* is used only for the next draw for the SVAR parameters).
*
compute sigmaAtBeta=SURGibbsSigma(bdraw)/tnobs
if draw<=0
next
*
* Do the bookkeeping here.
*
compute tgibbs(draw)=thetadraw
compute %modelsetcoeffs(nearvar,bdraw)
impulse(noprint,model=nearvar,factor=factor,steps=nsteps,$
flatten=%%responses(draw))
*
* Store the impulse responses
*
end do draw
infobox(action=remove)
*
@mcgraphirf(model=nearvar,$
shocklabels=shocklabels,varlabels=varlabels,$
percent=||.025,.16,.84,.975||,center=median,$
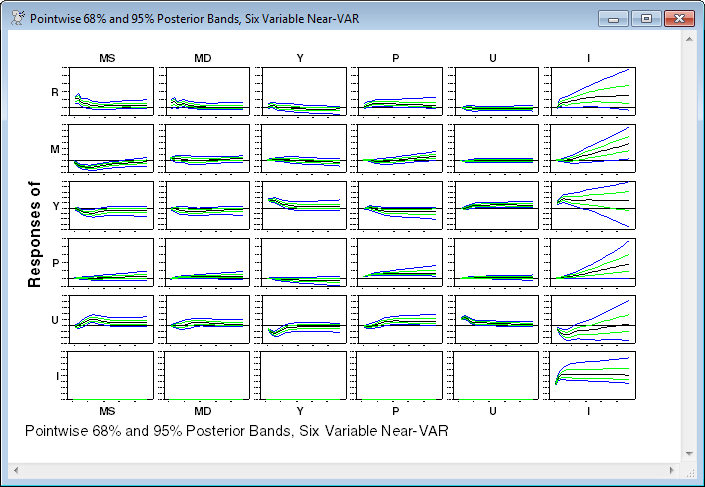
footer="Pointwise 68% and 95% Posterior Bands, Six Variable Near-VAR")
Output
The model is estimated using SUR since it doesn't have the same right-side variables in each equation.
Linear Systems - Estimation by Seemingly Unrelated Regressions
Iterations Taken 2
Quarterly Data From 1949:01 To 1989:03
Usable Observations 163
Log Likelihood 3590.3120
Dependent Variable FYGN3
Mean of Dependent Variable 0.0518561349
Std Error of Dependent Variable 0.0318224250
Standard Error of Estimate 0.0065462105
Sum of Squared Residuals 0.0069850182
Durbin-Watson Statistic 1.8425
Variable Coeff Std Error T-Stat Signif
*************************************************************************************
1. FYGN3{1} 1.134361652 0.080556800 14.08151 0.00000000
2. FYGN3{2} -0.639144328 0.129485544 -4.93603 0.00000080
3. FYGN3{3} 0.500220965 0.136348270 3.66870 0.00024379
4. FYGN3{4} -0.182773779 0.093808338 -1.94837 0.05137016
5. M1{1} 0.081932377 0.098342664 0.83313 0.40477054
6. M1{2} -0.224255124 0.163141065 -1.37461 0.16925279
7. M1{3} 0.303288259 0.152113094 1.99383 0.04617020
8. M1{4} -0.191669806 0.086888757 -2.20592 0.02738947
9. GNP82{1} 0.023662085 0.077602714 0.30491 0.76043233
10. GNP82{2} 0.040156577 0.099383392 0.40406 0.68617064
11. GNP82{3} -0.183734704 0.097935197 -1.87608 0.06064368
12. GNP82{4} 0.107979036 0.077350032 1.39598 0.16272076
13. GD{1} 0.123835892 0.104802738 1.18161 0.23736075
14. GD{2} 0.056931753 0.167766163 0.33935 0.73434471
15. GD{3} -0.156507372 0.164375202 -0.95213 0.34102855
16. GD{4} 0.015261957 0.101092923 0.15097 0.87999971
17. LHUR{1} 0.057724419 0.241062103 0.23946 0.81074991
18. LHUR{2} -0.028431465 0.373737652 -0.07607 0.93936076
19. LHUR{3} -0.204753887 0.379045986 -0.54018 0.58907140
20. LHUR{4} 0.030882172 0.237153047 0.13022 0.89639203
21. Constant 0.053493140 0.042626133 1.25494 0.20950139
22. GIN82{1} 0.068472294 0.027544549 2.48587 0.01292335
23. GIN82{2} -0.073264286 0.038683002 -1.89397 0.05822954
24. GIN82{3} 0.028652248 0.039015212 0.73439 0.46271316
25. GIN82{4} -0.011680214 0.026643826 -0.43838 0.66110828
Dependent Variable M1
Mean of Dependent Variable 5.4526945737
Std Error of Dependent Variable 0.5977479064
Standard Error of Estimate 0.0050988212
Sum of Squared Residuals 0.0042376704
Durbin-Watson Statistic 1.9320
Variable Coeff Std Error T-Stat Signif
*************************************************************************************
26. FYGN3{1} -0.634848867 0.064704306 -9.81154 0.00000000
27. FYGN3{2} 0.607372809 0.104004533 5.83987 0.00000001
28. FYGN3{3} -0.131023923 0.109516766 -1.19638 0.23154740
29. FYGN3{4} 0.018886109 0.075348120 0.25065 0.80208367
30. M1{1} 1.348249628 0.078990152 17.06858 0.00000000
31. M1{2} -0.213875817 0.131037100 -1.63218 0.10264208
32. M1{3} -0.178977827 0.122179285 -1.46488 0.14295403
33. M1{4} -0.013982105 0.069790220 -0.20034 0.84121096
34. GNP82{1} 0.002621955 0.062331545 0.04206 0.96644715
35. GNP82{2} 0.110083346 0.079826078 1.37904 0.16788246
36. GNP82{3} -0.079472337 0.078662869 -1.01029 0.31235620
37. GNP82{4} -0.049142585 0.062128587 -0.79098 0.42895458
38. GD{1} 0.034778698 0.084178970 0.41315 0.67949535
39. GD{2} -0.027236218 0.134752041 -0.20212 0.83982213
40. GD{3} 0.000598166 0.132028375 0.00453 0.99638513
41. GD{4} 0.054232469 0.081199197 0.66789 0.50420116
42. LHUR{1} -0.460004666 0.193624327 -2.37576 0.01751291
43. LHUR{2} 0.901261554 0.300191114 3.00229 0.00267955
44. LHUR{3} -0.493515747 0.304454839 -1.62098 0.10502155
45. LHUR{4} -0.066167753 0.190484521 -0.34737 0.72831674
46. Constant 0.048801088 0.034221253 1.42605 0.15385500
47. GIN82{1} -0.024391212 0.021880830 -1.11473 0.26496625
48. GIN82{2} 0.078614881 0.030532523 2.57479 0.01003005
49. GIN82{3} -0.078226568 0.030805347 -2.53938 0.01110483
50. GIN82{4} 0.055347179 0.021148990 2.61701 0.00887030
Dependent Variable GNP82
Mean of Dependent Variable 7.7256988534
Std Error of Dependent Variable 0.3655129025
Standard Error of Estimate 0.0086145804
Sum of Squared Residuals 0.0120963923
Durbin-Watson Statistic 1.7967
Variable Coeff Std Error T-Stat Signif
*************************************************************************************
51. FYGN3{1} -0.057773769 0.100577176 -0.57442 0.56568209
52. FYGN3{2} -0.248221974 0.161665935 -1.53540 0.12468546
53. FYGN3{3} 0.287662410 0.170234220 1.68980 0.09106552
54. FYGN3{4} -0.134807758 0.117122053 -1.15100 0.24973127
55. M1{1} 0.030469394 0.122783271 0.24816 0.80401378
56. M1{2} 0.123757295 0.203685693 0.60759 0.54345974
57. M1{3} -0.205421374 0.189916996 -1.08164 0.27941357
58. M1{4} 0.061462216 0.108482783 0.56656 0.57101184
59. GNP82{1} 0.982067830 0.096888927 10.13602 0.00000000
60. GNP82{2} 0.160262467 0.124082646 1.29158 0.19650318
61. GNP82{3} -0.163789959 0.122274539 -1.33953 0.18039940
62. GNP82{4} -0.000339649 0.096573446 -0.00352 0.99719384
63. GD{1} 0.115104542 0.130848835 0.87968 0.37903501
64. GD{2} -0.040533410 0.209460244 -0.19351 0.84655674
65. GD{3} -0.256257580 0.205226544 -1.24866 0.21179052
66. GD{4} 0.164787529 0.126217038 1.30559 0.19169249
67. LHUR{1} -0.214124501 0.300972055 -0.71144 0.47680968
68. LHUR{2} 0.746530957 0.466620790 1.59987 0.10962821
69. LHUR{3} -0.202784257 0.473248378 -0.42849 0.66829124
70. LHUR{4} -0.047325036 0.296091501 -0.15983 0.87301305
71. Constant 0.042104854 0.053267854 0.79044 0.42927292
72. GIN82{1} 0.060396904 0.035082906 1.72155 0.08515145
73. GIN82{2} -0.042876627 0.049815780 -0.86070 0.38940124
74. GIN82{3} -0.020920922 0.050214348 -0.41663 0.67694735
75. GIN82{4} 0.027452270 0.033981395 0.80786 0.41917017
Dependent Variable GD
Mean of Dependent Variable 3.8601005333
Std Error of Dependent Variable 0.5480266865
Standard Error of Estimate 0.0047092303
Sum of Squared Residuals 0.0036148265
Durbin-Watson Statistic 2.0535
Variable Coeff Std Error T-Stat Signif
*************************************************************************************
76. FYGN3{1} 0.189489949 0.060050494 3.15551 0.00160218
77. FYGN3{2} -0.118281110 0.096524079 -1.22541 0.22042258
78. FYGN3{3} 0.075997918 0.101639850 0.74772 0.45463043
79. FYGN3{4} -0.091101140 0.069928760 -1.30277 0.19265305
80. M1{1} 0.160237248 0.073308840 2.18578 0.02883145
81. M1{2} -0.164956080 0.121612348 -1.35641 0.17496908
82. M1{3} 0.044424698 0.113391625 0.39178 0.69521994
83. M1{4} -0.019324861 0.064770607 -0.29836 0.76542957
84. GNP82{1} 0.007044415 0.057848392 0.12177 0.90307821
85. GNP82{2} -0.028231042 0.074084644 -0.38106 0.70315525
86. GNP82{3} -0.071986175 0.073005097 -0.98604 0.32411195
87. GNP82{4} 0.088495861 0.057660032 1.53479 0.12483619
88. GD{1} 1.295165907 0.078124457 16.57824 0.00000000
89. GD{2} -0.108044540 0.125060095 -0.86394 0.38762032
90. GD{3} -0.162590521 0.122532327 -1.32692 0.18453535
91. GD{4} -0.059048232 0.075359001 -0.78356 0.43329886
92. LHUR{1} 0.214171361 0.179698033 1.19184 0.23332386
93. LHUR{2} -0.097546946 0.278600078 -0.35013 0.72623925
94. LHUR{3} 0.033575863 0.282557138 0.11883 0.90541117
95. LHUR{4} 0.009823515 0.176784055 0.05557 0.95568605
96. Constant -0.022334201 0.031757567 -0.70327 0.48188638
97. GIN82{1} -0.004014643 0.020272521 -0.19803 0.84301869
98. GIN82{2} 0.052905855 0.028259797 1.87212 0.06118940
99. GIN82{3} -0.029620898 0.028513868 -1.03882 0.29888647
100. GIN82{4} -0.006272017 0.019592114 -0.32013 0.74887002
Dependent Variable LHUR
Mean of Dependent Variable 0.0570695297
Std Error of Dependent Variable 0.0167072122
Standard Error of Estimate 0.0027801361
Sum of Squared Residuals 0.0012598526
Durbin-Watson Statistic 1.8212
Variable Coeff Std Error T-Stat Signif
*************************************************************************************
101. FYGN3{1} -0.006146236 0.032825868 -0.18724 0.85147435
102. FYGN3{2} 0.045972472 0.052763707 0.87129 0.38359602
103. FYGN3{3} -0.093616737 0.055560180 -1.68496 0.09199609
104. FYGN3{4} 0.083200414 0.038225701 2.17656 0.02951364
105. M1{1} -0.036394556 0.040073380 -0.90820 0.36377373
106. M1{2} -0.010453758 0.066477902 -0.15725 0.87504654
107. M1{3} 0.024413706 0.061984144 0.39387 0.69367689
108. M1{4} 0.015103243 0.035406060 0.42657 0.66969091
109. GNP82{1} -0.106195423 0.031622116 -3.35826 0.00078434
110. GNP82{2} 0.017278016 0.040497464 0.42664 0.66963835
111. GNP82{3} 0.059906917 0.039907343 1.50115 0.13331671
112. GNP82{4} 0.027645563 0.031519151 0.87710 0.38043032
113. GD{1} 0.002080696 0.042705778 0.04872 0.96114113
114. GD{2} -0.017574881 0.068362570 -0.25708 0.79711440
115. GD{3} 0.073267302 0.066980797 1.09386 0.27401843
116. GD{4} -0.048423168 0.041194076 -1.17549 0.23979928
117. LHUR{1} 1.174134997 0.098229730 11.95295 0.00000000
118. LHUR{2} -0.400219023 0.152293323 -2.62795 0.00859015
119. LHUR{3} -0.041766146 0.154456401 -0.27041 0.78684689
120. LHUR{4} 0.172371521 0.096636840 1.78370 0.07447177
121. Constant 0.022203504 0.017381799 1.27740 0.20146119
122. GIN82{1} -0.001911290 0.011400202 -0.16765 0.86685548
123. GIN82{2} 0.003227095 0.016149507 0.19983 0.84161648
124. GIN82{3} -0.007717547 0.016280729 -0.47403 0.63547881
125. GIN82{4} 0.006209713 0.011039059 0.56252 0.57376047
Dependent Variable GIN82
Mean of Dependent Variable 5.4930861728
Std Error of Dependent Variable 0.4432821202
Standard Error of Estimate 0.0259108781
Sum of Squared Residuals 0.1094338972
Durbin-Watson Statistic 1.9376
Variable Coeff Std Error T-Stat Signif
*************************************************************************************
126. Constant 0.009838916 0.025471195 0.38628 0.69929213
127. GIN82{1} 1.446070599 0.078112684 18.51262 0.00000000
128. GIN82{2} -0.464795005 0.137450841 -3.38154 0.00072082
129. GIN82{3} -0.007309488 0.137281274 -0.05324 0.95753701
130. GIN82{4} 0.025152776 0.078119689 0.32198 0.74746979
Covariance\Correlation Matrix of Residuals
FYGN3 M1 GNP82 GD LHUR GIN82
FYGN3 4.2853e-005 0.06777237 0.21129919 0.10058751 -0.37757048 0.32853059
M1 2.2621e-006 2.5998e-005 0.22777114 0.17783435 -0.21474695 0.16136926
GNP82 1.1916e-005 1.0005e-005 7.4211e-005 0.01139069 -0.63047513 0.48672522
GD 3.1009e-006 4.2701e-006 4.6210e-007 2.2177e-005 -0.15290033 0.11257864
LHUR -6.8715e-006 -3.0441e-006 -1.5100e-005 -2.0018e-006 7.7292e-006 -0.45875454
GIN82 5.5725e-005 2.1319e-005 1.0864e-004 1.3737e-005 -3.3047e-005 6.7137e-004
## NL6. NONLIN Parameter A12 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A21 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A23 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A24 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A31 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A36 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A41 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A43 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A46 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A51 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A53 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A54 Has Not Been Initialized. Trying 0
## NL6. NONLIN Parameter A56 Has Not Been Initialized. Trying 0
Covariance Model-Marginal Posterior - Estimation by BFGS
Convergence in 29 Iterations. Final criterion was 0.0000069 <= 0.0000100
Observations 163
Function Value 3856.6130
Prior DF 3.5000
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. A12 -0.631672606 0.368867847 -1.71246 0.08681134
2. A21 0.348463823 0.214584410 1.62390 0.10439695
3. A23 -0.201732085 0.080765034 -2.49777 0.01249790
4. A24 -0.245472883 0.118384690 -2.07352 0.03812402
5. A31 0.066098765 0.145360003 0.45472 0.64930740
6. A36 -0.167307846 0.026854301 -6.23021 0.00000000
7. A41 0.015087985 0.079542147 0.18969 0.84955565
8. A43 0.027267747 0.051141542 0.53318 0.59390763
9. A46 -0.026125504 0.016792250 -1.55581 0.11975394
10. A51 0.091762255 0.024822056 3.69680 0.00021833
11. A53 0.169559780 0.020426281 8.30106 0.00000000
12. A54 0.065962313 0.032983812 1.99984 0.04551765
13. A56 0.012818208 0.007155073 1.79149 0.07321544
Graph
The graph is probably too "busy" to be displayed in a 6 x 6 matrix (certainly at this size). Two things to note: the zero response of investment to the first five shocks is by construction. This is due to a combination of the other variables being excluded from the lags in the investment equation and the other shocks being excluded from the definition of the investment shock in the structural model. The other is that the misspecification of the dynamics of investment (which is treated as exogenous for illustration) causes it to be borderline unstable, and thus its shocks dominate all the responses.
Copyright © 2026 Thomas A. Doan